I am a highly motivated Computational Biologist and recently graduated from UCL with practical experience
of big data analysis, bioinformatics, image processing, software engineering, machine learning
(ML) and deep learning (AI) for image segmentation/classification, and advanced mathematical modelling
of biological phenomena.
I am taking a gap year before pursuing a PhD in computational biology or related discipline.
My ambition is to employ my pertinent analytical, software, and communication skills to
create data-driven improvements for human health. I am passionate about big data in biology, as
we begin to answer some of the most fundamental and exciting questions about living systems.
EDUCATION
2019—2023 MSci Biological Sciences: Computational Biology University College London (UCL)Fist Class Honours | Ranked second among 39 Biological Sciences finalists
Obtained strong analytic, software, and communication skills in Biology and Pharmaceutical contexts through the following modules:
- Research software engineering with Python
- Dynamic biological systems, Understanding bioinformatics resources and their application
- Advanced computational biology
- Advanced molecular cell biology
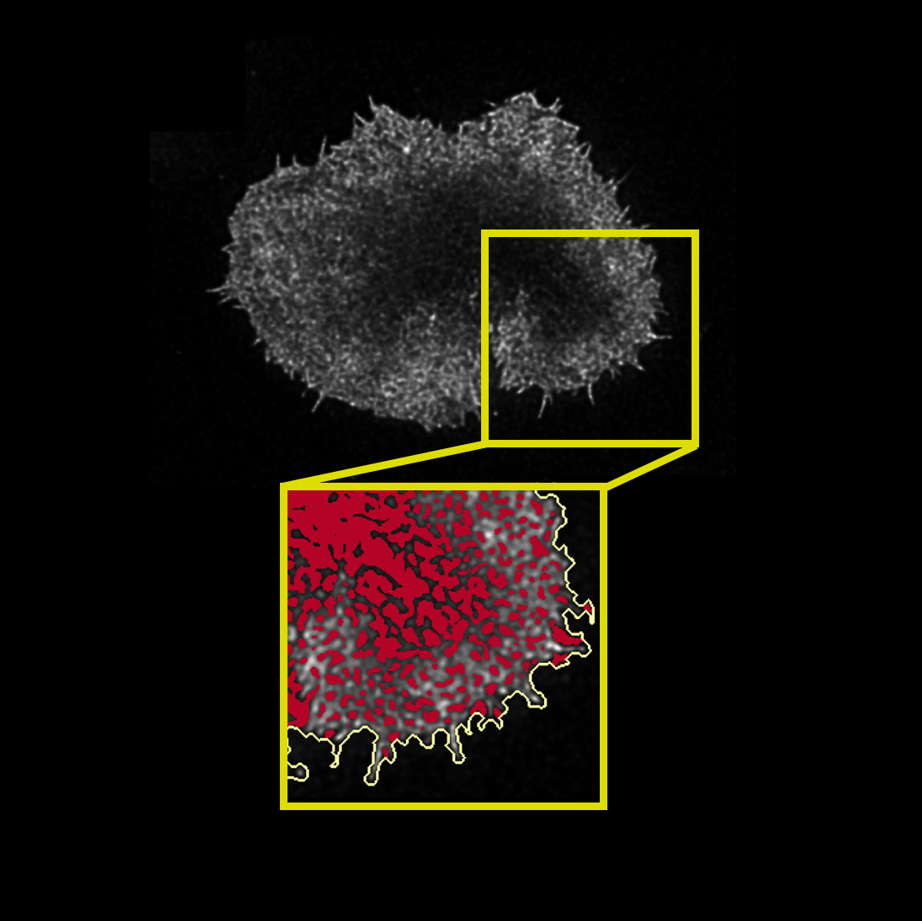
4th year MSci Extended Research Project (45 ECTS): Cytoskeletal actin remodelling at the immunological synapse: A quantitative comparison between T and CAR-T cells.
Host group: Simoncelli group at the London Centre for Nanotechnology, UCL.
Quantified cytoskeletal actin dynamics in cancer immunotherapy T cells from microscopy data; implemented an installable, semi-automated, documented Python pipeline/library (see GitHub).
- Data-lead interdisciplinary problem solving: using best practices in software engineering, created a documented tool; extracted biologically-relevant parameters from ~100 super-resolution microscopy images.
- Working in a cross-functional, interdisciplinary team: being proactive, innovative, and pragmatic, conducted an unprecedented comparison between primary and bioengineered cancer immunotherapy cells.
Host group: Lane Origins group, Genetics, Evolution, and Environment, UCL.
Implemented a stochastic kinetic model of protometabolic reactions in R; reviewed suitable mathematical modelling methods, extensions, and advanced frequentist and Bayesian parameter space exploration techniques (see GitHub).
- Adaptable, innovative and uninhibited problem-solving: research on the origin of life requires a very rigorous but also unhinged, daring, and creative approach to unique challenging problems.
2017—2019 International Baccalaureate (IB) Diploma United World College of the Adriatic
Bilingual diploma awarded with 43/45 points.
- Higher Level subjects: Chemistry (7), Biology (6), Russian Literature (7).
- Standard Level subjects: Mathematics (7), Global Politics (6), English B (7), Italian B (6) (certificate).
TECHNICAL SKILLS
Programming languages (example libraries):- Advanced: Python (numpy, scipy, pandas, scikit-learn, seaborn, openCV, pytest, requests, TensorFlow), R/RStudio (dplyr, tidyrtidyverse, ggplot2, shiny, pomp, GillespieSSA).
- Intermediate: MATLAB (Image Processing Toolbox), bash, SQL.
Analytical skills: frequentist and Bayesian statistical analysis, supervised machine learning, convolutional neural networks (CNNs), predictive modelling, big data analysis.
Software engineering: agile development, version control, testing, deployment, continuous integration.
LANGUAGE SKILLS
| Native | C2 | B2-C1 |
|---|---|---|
| Turkish, Bulgarian | Russian, English | Italian |
PROFESSIONAL EXPERIENCE (TECHNICAL)
Data scientist (epidemiological modelling) at the Big Data Institute, University of Oxford (UK) Aug, 2023 - present-
Assisting Prof Hollingsworth and an interdisciplinary team of researchers and consultants
working on an externally commissioned, policy-facing project to evaluate the health and
economic impacts of transmission and vaccination scenarios for neglected tropical diseases.
- Used R and Stan for Bayesian estimation to infer the time-varying force of infection from a large curated seroprevalence data set.
- Collaboratively set up a multilevel transmission metapopulation model to evaluate various transmission and vaccination scenarios for Chikungunya virus using geospatial data.
- Devised creative visualisations and delivered updates at external working group presentations.
Computational researcher (summer intern) at the Randall Centre for Cell & Molecular Biophysics, King's College London (UK) Jun-Aug, 2022
-
Developed a Python tool to extract biologically-relevant information from microscopy images
of yeast microtubules, exploring applied mathematics and deep learning approaches.
- Funded by one of three Life Sciences undergraduate summer studentships offered by the Royal Microscopical Society (RMS).
- Explored and implemented several different techniques of detecting lines in microscopy images and of quantifying biologically relevant parameters using Python (see GitHub), ranging from mathematical parametrisation to convolutional neural networks (CNNs).
- Presented work to two other groups at the institute and published a report on the summer studentship in the infocus member magazine of the Royal Microscopical Society.
Data scientist (summer intern) at the Oxford Big Data Institute (remote / Oxford, UK) Jul-Sep, 2021
-
Used advanced stochastic modelling to understand the impacts of waning vaccination
immunity against SARS-CoV-2; characterised transmission using multiple data sources.
- Showed adaptability and perseverance while exploring advanced deterministic and stochastic modelling techniques and R packages to model waning infection and vaccination immunity against SARS-CoV-2 (see GitHub); remained curious, driven, and flexible in the face of logistic and conceptual challenges.
- Digested complex problems and developed analytical solutions by processing, transforming, and integrating data from public repositories and peer-reviewed publications for further visualisation and analysis; tackled problems of combining large data sets from multiple sources(see GitHub).
- Completed the ONS safe researcher training course, a GDPR-compliant data privacy and security training to access ONS databases (Office for National Statistics).
PROFESSIONAL EXPERIENCE (TRANSFERABLE)
Online private tutor at Lanterna Education (remote, UK) Dec 2019-present- Developed strong communication skills by having delivered 250+ hours of Maths, Chemistry, Biology (SL and HL) tutoring.
- Built strong rapport with all stakeholders, as evidenced by retention bonuses received for ~1/3 of students tutored rebooking tuition with me because they appreciated my individual approach, the accessible material delivery, while the parents valued my reliability.
Transition mentor at UCL Transitions Programme (remote, UK) Sep 2020 - Mar 2021 and Sep-Dec, 2022
- Senior Transition mentor: exhibited excellent team-building and organisations skills; organised ~90 students into 10 mentoring groups, monitored weekly attendance, responded to queries and offered individualised support to mentors and mentees alike.
- Junior transition mentor: showed independent work and collaboration skills; worked independently and in a team to structure and deliver weekly sessions to help first-year students transition into the Biological Sciences degree.
Sustainability and inclusivity intern at UCL EAST (remote/London, UK) Sep 2020 - Mar 2021 and Sep-Dec, 2022
- Promoted diversity, equality, and sustainability at the new UCL EAST campus by designing and conducting surveys to integrate staff
- Planned and developed a resource pack with interactive activities about sustainability and inclusivity to be used by future UCL Summer Schools.
- Worded in a cross-functional teams and integrated inputs and feedback from different departments of UCL/UCL EAST in line with promoting the UCL Ways of Working.
PUBLICATIONS AND AWARDS
- Altenhoff AM, Train CM, Gilbert KJ, Mediratta I, Mendes de Farias T, Moi D, Nevers Y, Radoykova HS, Rossier V, Warwick Vesztrocy A, Glover NM, Dessimoz C. OMA orthology in 2021: website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Res. 2021 Jan 8;49(D1):D373-D379. doi: 10.1093/nar/gkaa1007.
- Culley S and Radoykova S. Using image processing to quantify biologically relevant information in microscopy imagesUsing image processing to quantify biologically relevant information in microscopy images. infocus Magazine. 2023 Mar 6, Issue 69.
- Harold and Olga Fox Award for best 3rd year MSci Investigative Project symposium presentation. 2022 Jul. Project title: Modelling the kinetics of protometabolism.