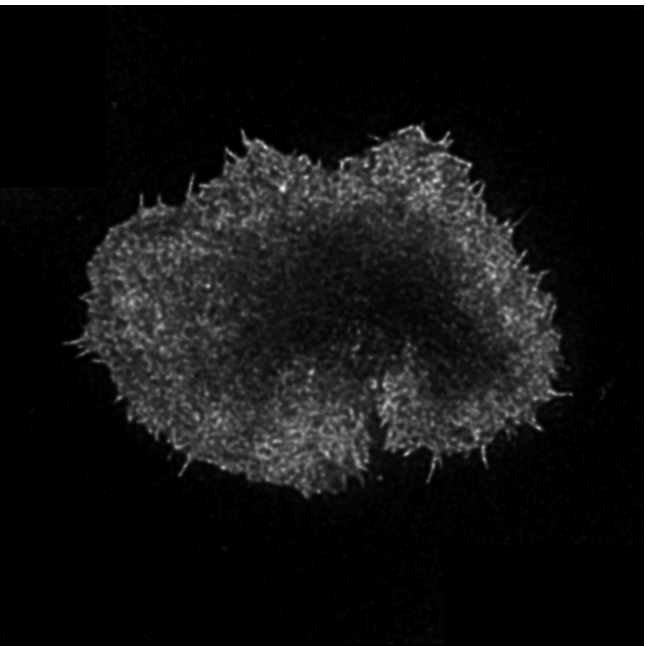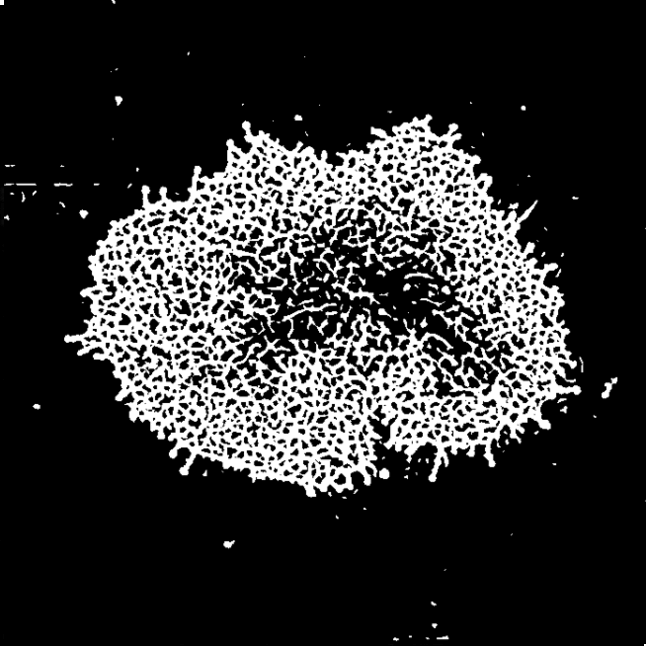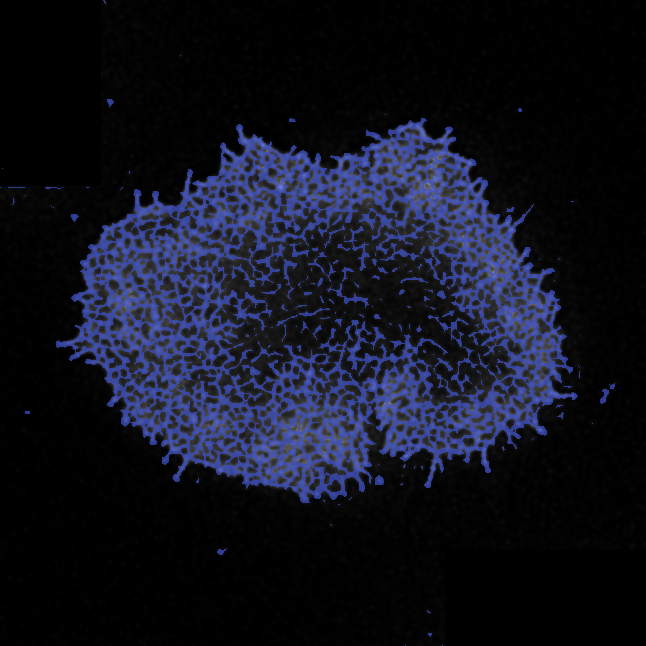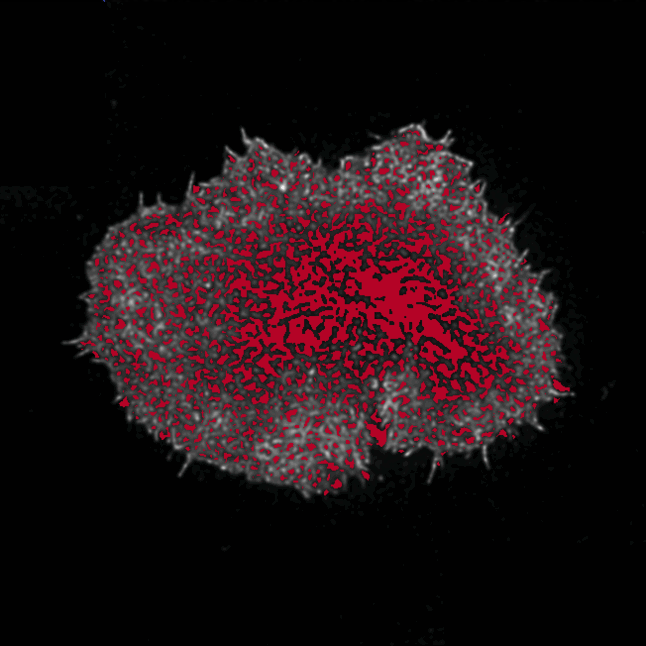
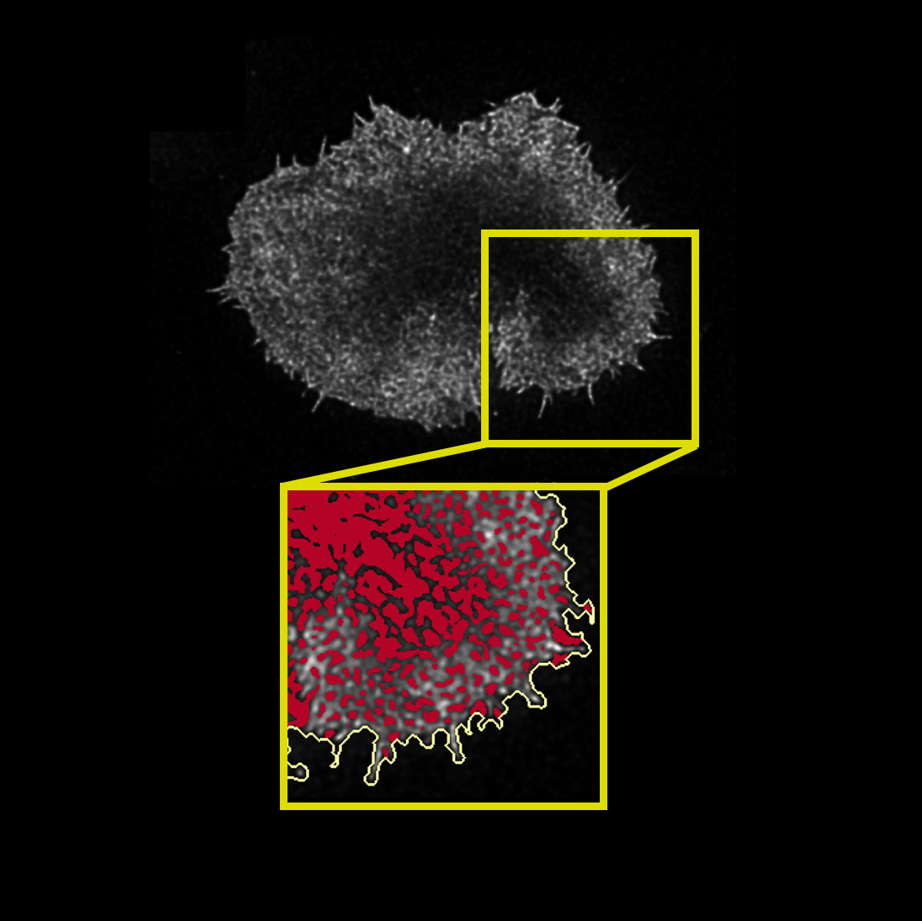
Aim: ActinMeshure is an open-source Python library for image segmentation. It was developed
for super-resolution microscopy images of cytoskeletal actin meshwork but can be applied to a broader range
of segmentation tasks.
Methods: implemented state of the art image processing and edge detection algorithms,
morphological operations, using libraries such as numpy, sci-kit image, scipy, openCV
to segment the mesh structure. Implemented classes for batch-processing and tools for interactive analysis with
semi-automated hyperparameter tuning.
Output: An installable and documented open-source Python library for biomedical image processing.
Key words: Python, software engineering, image processing/segmentation tool, super-resolution microscopy.
Aim: extract biologically relevant quantitative information from microscopy images
of S. cerevisiae (baker's yeast) cells using applied mathematics and/or deep learning methods.
Methods: curated data using ImageJ macros; implemented the linear Hough transform in Python;
used Gaussian fitting (SciPy, sci-kit learn) and FWHM for line curation; compared the Hough
transform to linear regression; hyperparameter tuning for a proposed specific U-net architecture to train a CNN.
Output: repo published on group's GitHub presented work at joint group meeting; wrote a concise two-page report, published in the
infocus magazine.
Key words: Python, ImageJ macros, image processing/segmentation, applied mathematics.
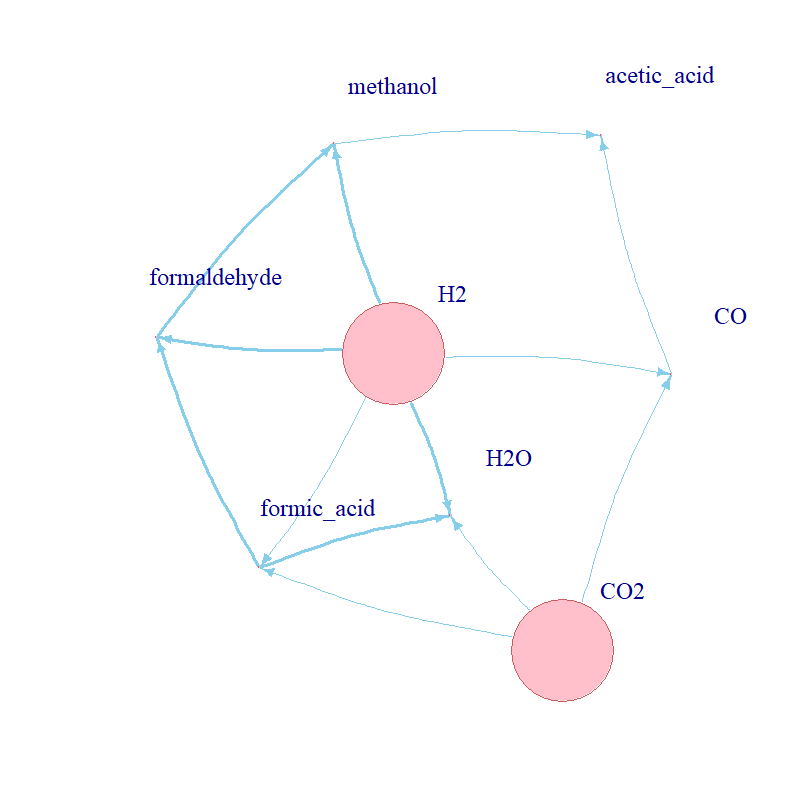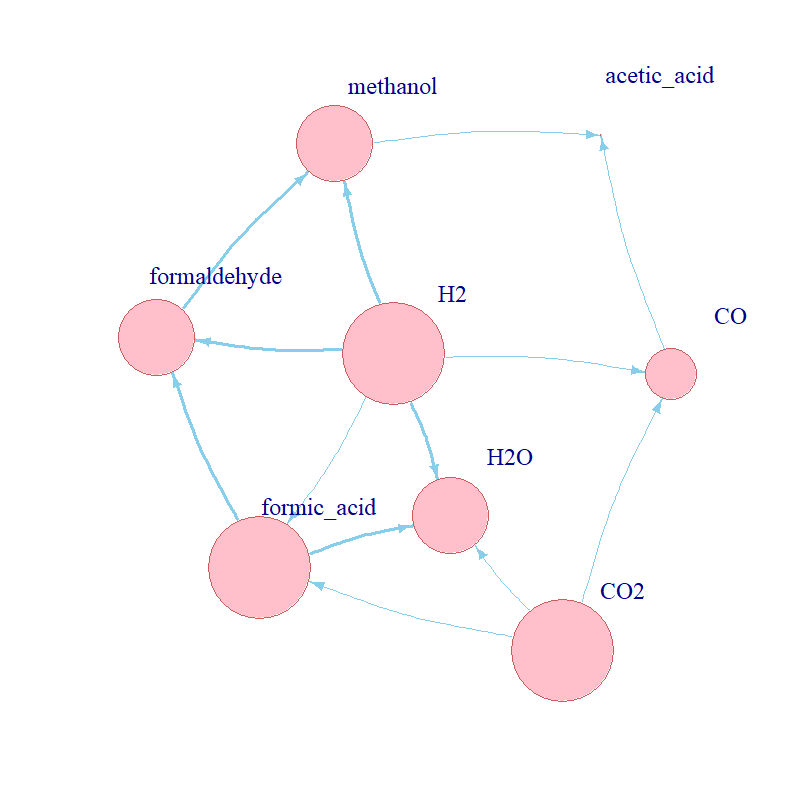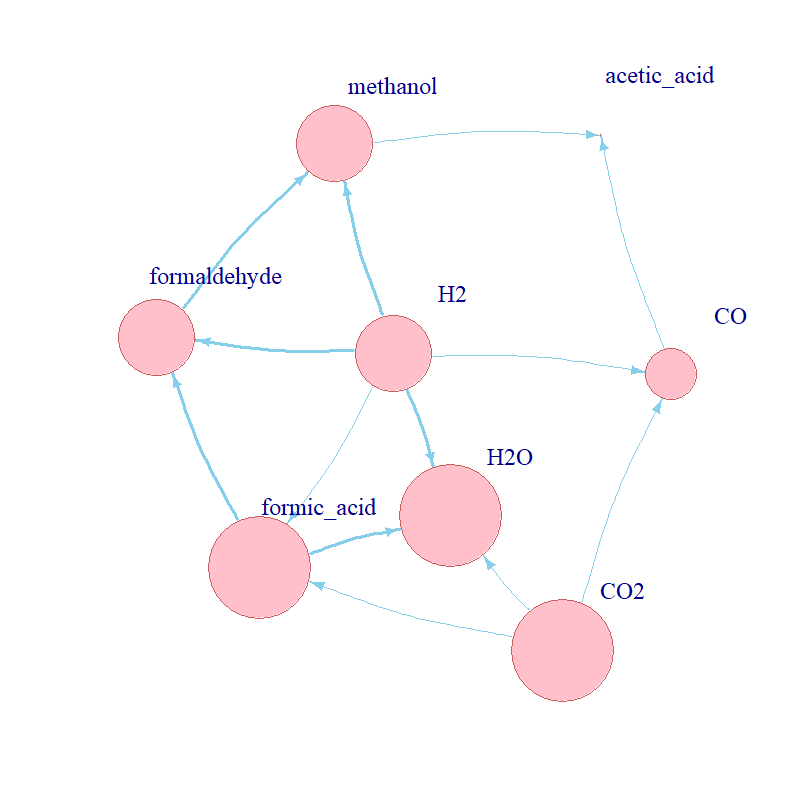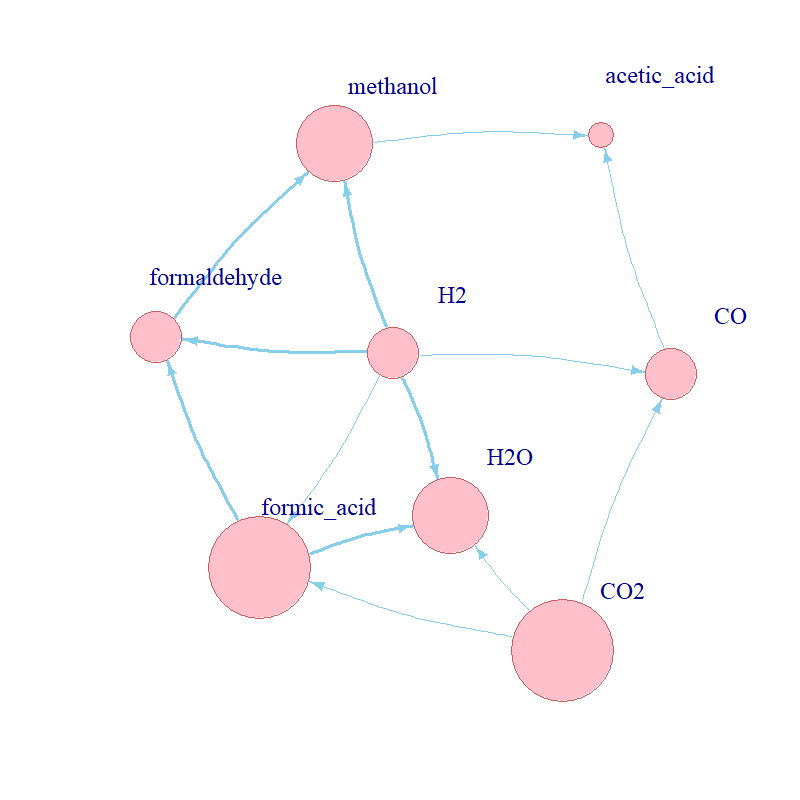
Aim: understand the kinetic conditions which could enable and/or drive the evolution of
metabolism at the origin of life.
Methods: implemented a stochastic kinetic model of a curated set of biochemical reactions in R
to explore the behaviour of a proposed protometabolic network; simulated the dynamics of the network using the Gillespie
algorithm; reviewed advanced mathematical modelling and parameter space exploration techniques for expansion of the model;
conducted a brute-force parameter scan.
Output: 7,000 word report with a literature review component; was awarded Harold and Olga Fox
prize for best MSci Investigative project symposium presentation.
Key words: stochastic modelling, Gillespie algorithm, R/RStudio, parameter space exploration, dimensionality curse.
Aim: disease transmission is often modelled using the negative binomial distribution. This project
integrated data from multiple sources with the aim of understanding whether this assumption held for SARS-CoV-2 transmission.
Methods: integrated large datasets from multiple sources by mapping relevant variables; creatively visualised the data to spot
patterns in viral transmissibility.
Output: creative visualisations which highlight age-stratified differences in SARS-CoV-2 transmission; presented
work at GEE Undergraduate Symposium (Sep. 2021).
Key words: R/RStudio (ggplot2), Python (matplotlib, seaborn), transmission matrix,
mathematical modelling.
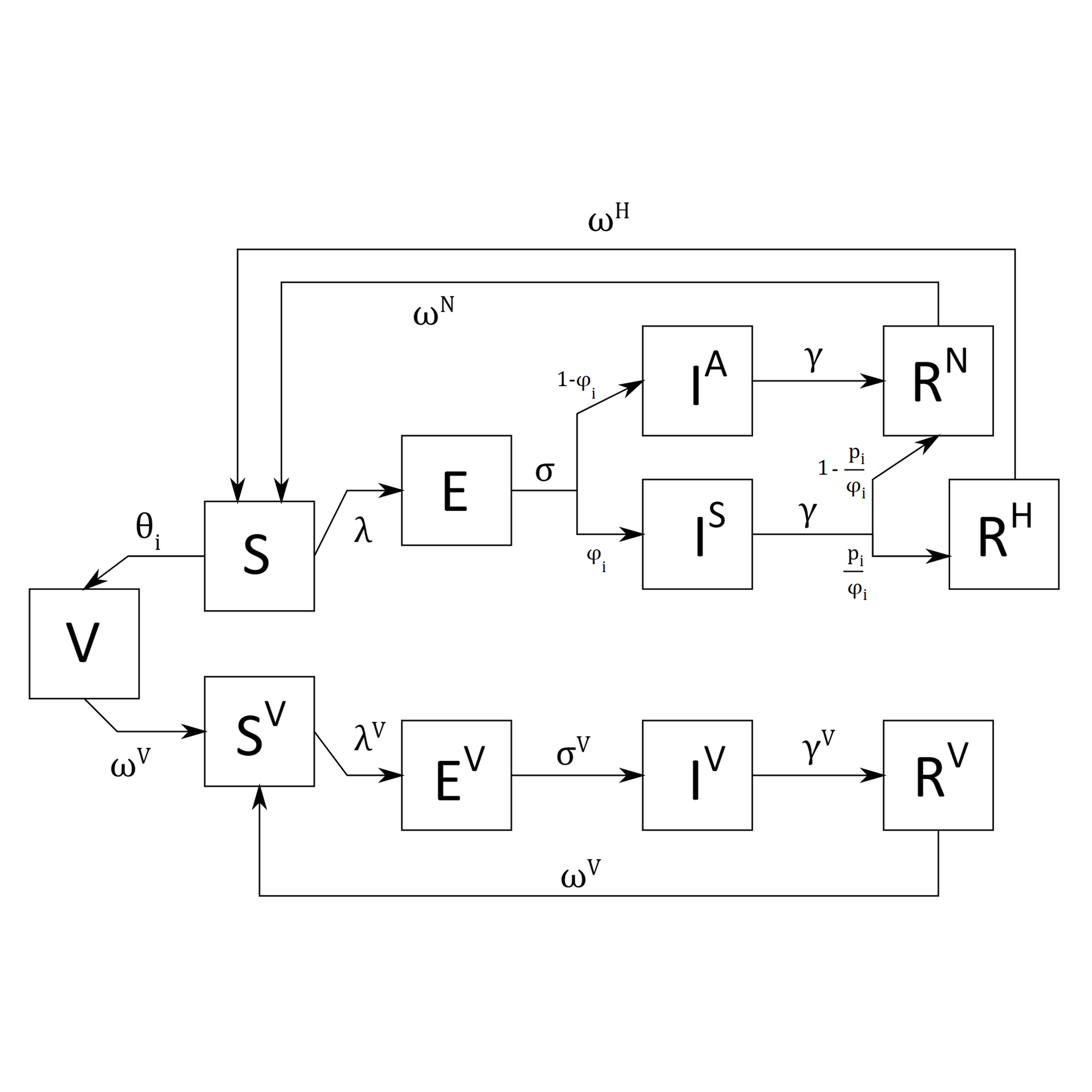
Aim: expanding a difference equation transmission-dynamic model of COVID-19 to include the effects of
vaccination and waning vaccination immunity.
Methods: introduced compartments and difference equations for vaccination and waning immunity;
explored the possibility of using Partially Observed Markov Process (POMP) models for more detailed description
of the transmission dynamics.
Output: template for the simulation in R/RStudio and a report outlining the new equations.
Key words: partially observed Markov process, dynamic modelling, age-structured deterministic model.